Replace a failed hard drive in a RAID array
One of our servers started beeping very loudly and disturbing the people working in the same room. What happened? It turns out that the RAID device was beeping due to a failed hard drive. In this post, we will walk through all the steps to find evidence of the disk failure, identify the drive, replace it, and rebuild the array.
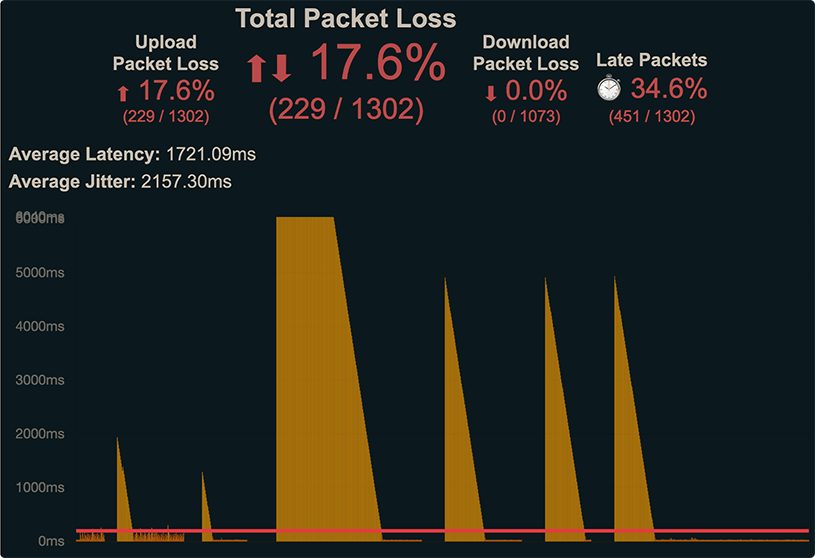
Reduce packet loss on a cable internet connection by increasing power
An internet connection with high packet loss is frustating to use. When packets of data are lost, phone calls over wifi can be randomly punctuated with silence. Zoom meetings might get frozen and unfrozen at unpredictable times. Using the internet is unpleasant when the connection is unstable. Here, I will show that one reason for high packet loss can be low power in the coaxial cable that connects to the modem.
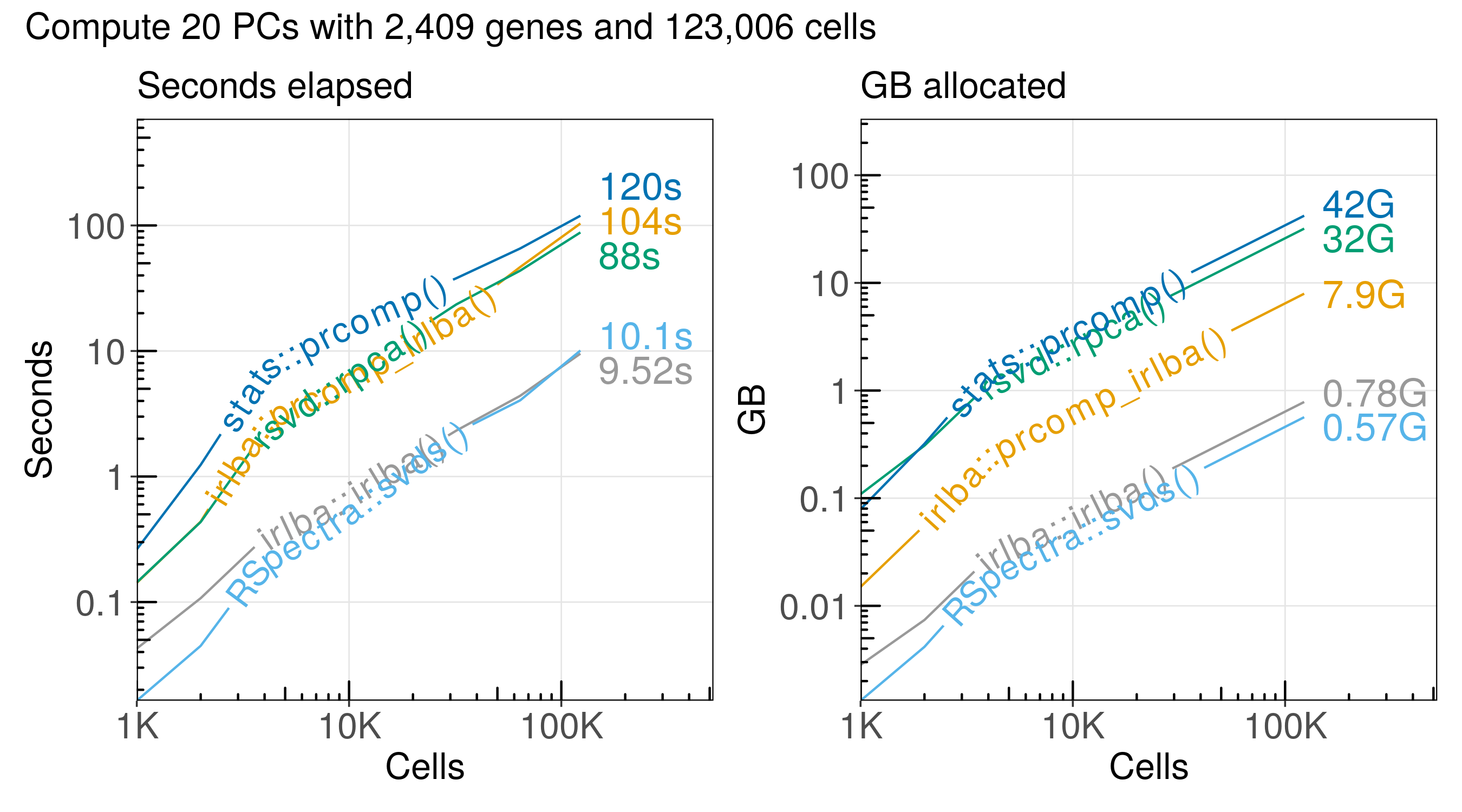
Benchmark principal component analysis (PCA) of scRNA-seq data in R
Principal component analysis (PCA) is frequently used for analysis of single-cell RNA-seq (scRNA-seq) data. We can use it to reduce the dimensionality of a large matrix with thousands of features (genes) to a smaller matrix with just a few factors (principal components). Since the latest scRNA-seq datasets include millions of cells, there is a need for efficient algorithms. Specifically, we need algorithms that work with sparse matrices instead of dense matrices. Here, we benchmark five implementations of singular value decomposition (SVD) and PCA.
Debug a workflow on the Terra platform
Some biomedical researchers use the Terra platform to run data analysis jobs on Google Cloud. When we run into errors, it can be daunting to figure out where the errors are coming from and how to fix them. In this tutorial, we walk through each step of viewing an error, understanding it, and fixing it in an actual workflow. I hope this illustrates the general strategy for how we can solve any workflow issue on Terra.
Tools for exploring the scientific literature
There are millions of scientific publications, and many people have created tools for exploring them. In this post, we’ll highlight a few websites and tools that allow us to search for text inside figures, view citation networks, share public comments, and more.
Find the most abundant barcodes in FASTQ files
Single-cell RNA-seq data contains oligonucleotide barcodes to uniquely identify each multiplexed sample, each single cell, and each individual molecule. Can we check which barcodes are present in a given FASTQ file? Maybe we can guess which 10x sample index was used during library preparation?
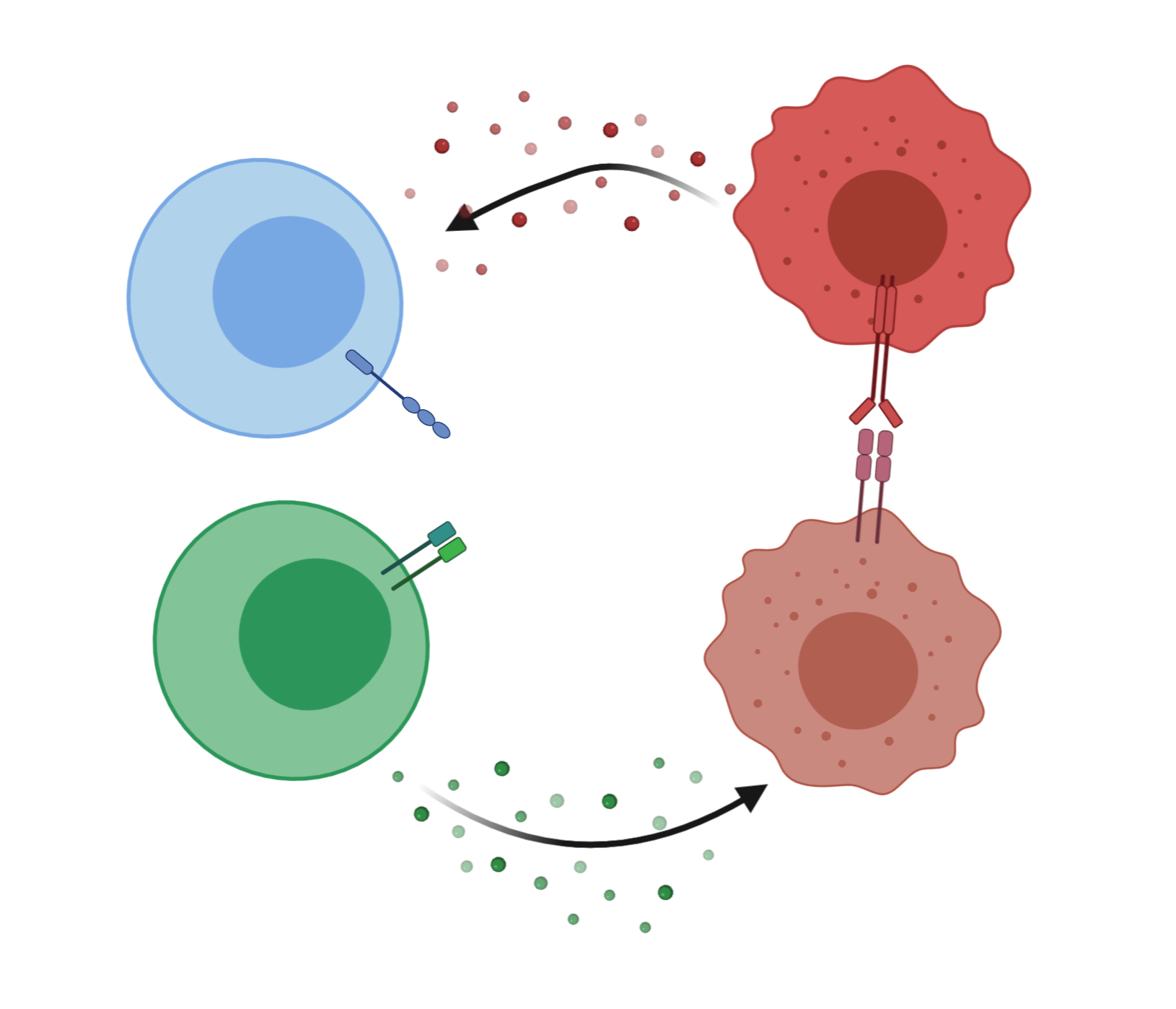
Make a table with ligands and receptors in R with OmnipathR
Curated lists of genes help computational biologists to focus analyses on a subset of genes that might be important for a research question. For example, we might be interested to focus on the genes encoding the signals and receptors for cell-to-cell communication. OmnipathR is a new R package that provides access to a vast database of genes called OmniPath, organized and curated by the Saez-Rodriguez Lab. Let’s try to use OmnipathR to create a simple table with ligands and receptors.
Resources for illustrating biomedical science
Clear illustrations are essential for effective science communication. Here, we’ll mention a few resources that might be of interest for researchers, educators, or anyone interested in making figures and presentations about biomedical science.
Make a table with your most recent coauthors in R
Some grant agencies might require a table that lists all of your coauthors, departments, and dates for publications from the last few years. Making such a table can be a laborious task for academics who publish papers with a lot of coauthors. Here, we’ll download our publication records from NCBI and use R to automatically make a table of coauthors.
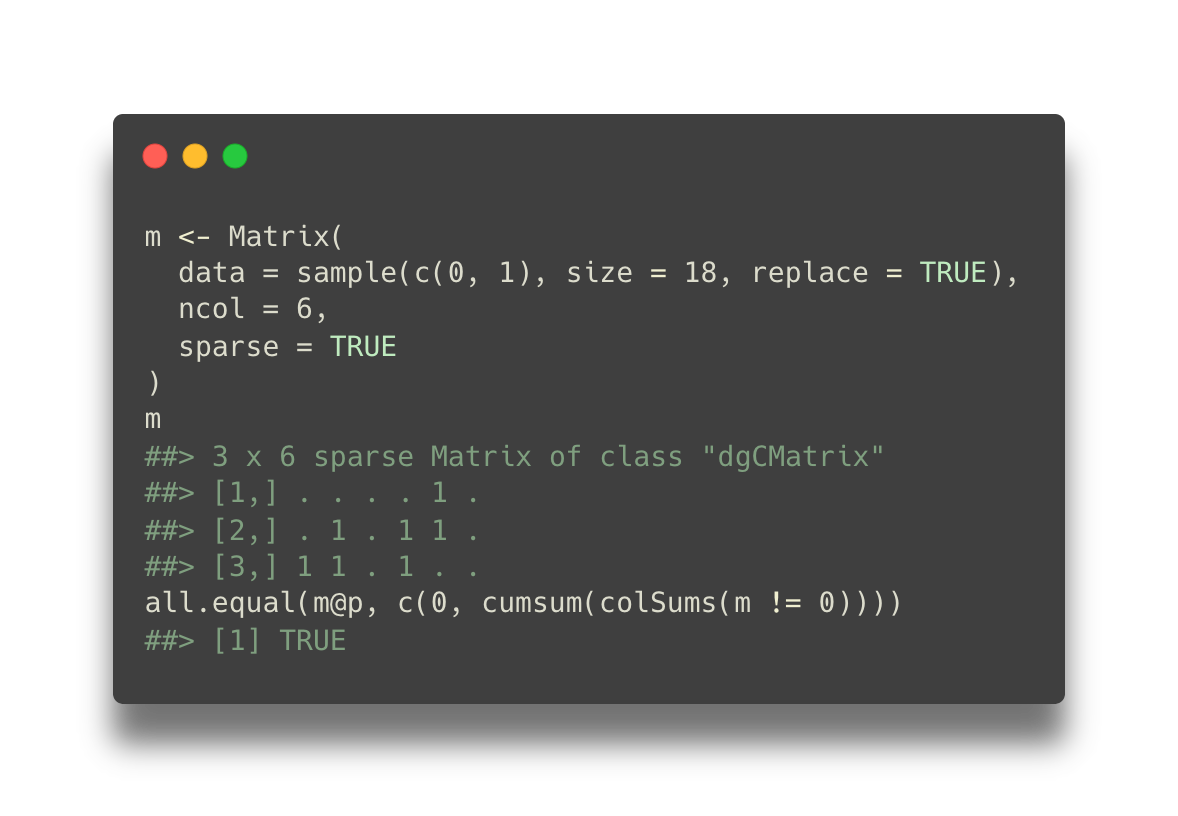
Working with a sparse matrix in R
Sparse matrices are necessary for dealing with large single-cell RNA-seq
datasets. They require less memory than dense matrices, and they allow some
computations to be more efficient. In this note, we’ll discuss the internals of
the dgCMatrix class with examples.
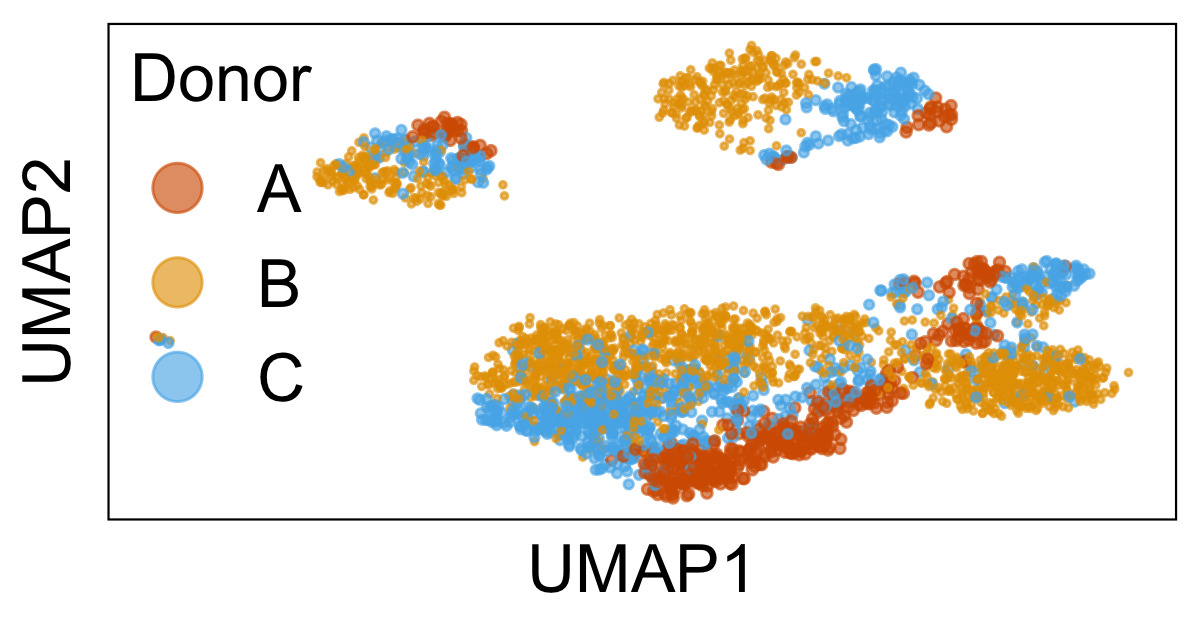
Harmony in motion: visualize an iterative algorithm for aligning multiple datasets
Harmony is a an algorithm for aligning multiple high-dimensional datasets, described by Ilya Korsunsky et al. in this paper. When analyzing multiple single-cell RNA-seq datasets, we often encounter the problem that each dataset is separated from the others in low dimensional space – even when we know that all of the datasets have similar cell types. To address this problem, the Harmony algorithm iteratively clusters and adjusts high dimensional datasets to integrate them on a single manifold. In this note, we will create animated visualizations to see how the algorithm works and develop an intuitive understanding.
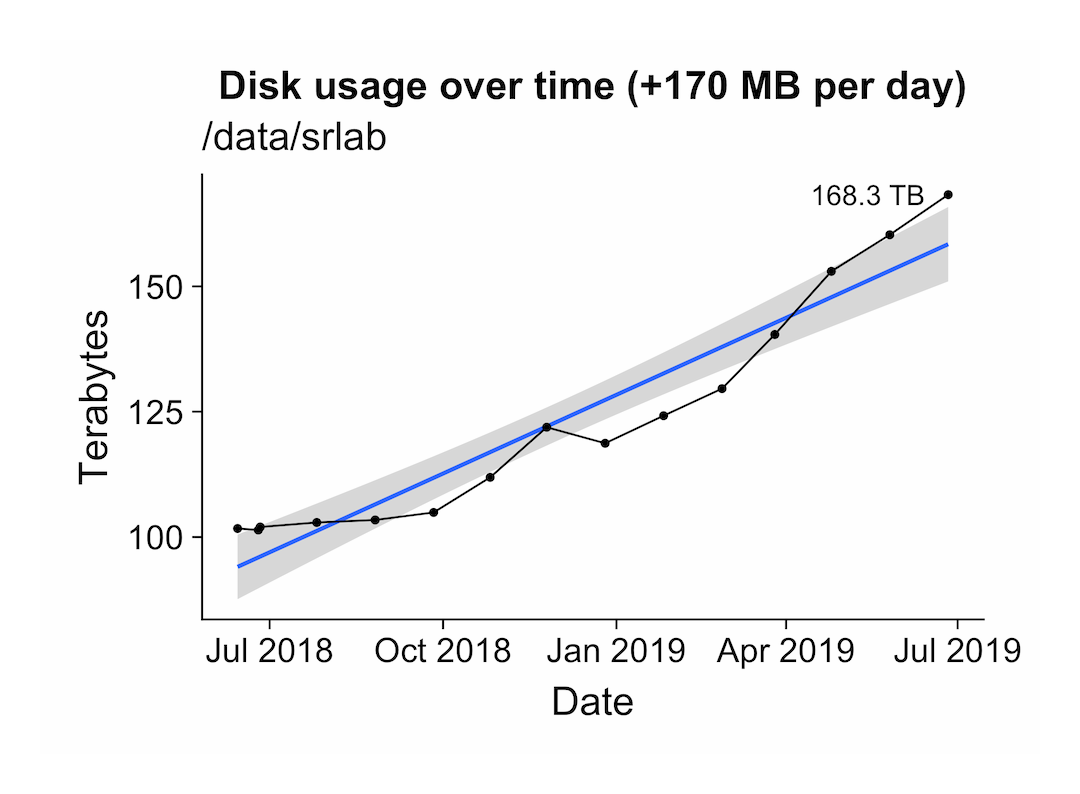
Monitor disk usage on your server
You might consider monitoring your disk usage, because this might reveal trends that help you to plan for the future. Here, we’ll use a cron job to periodically scan a directory with the duc program by Ico Doornekamp.
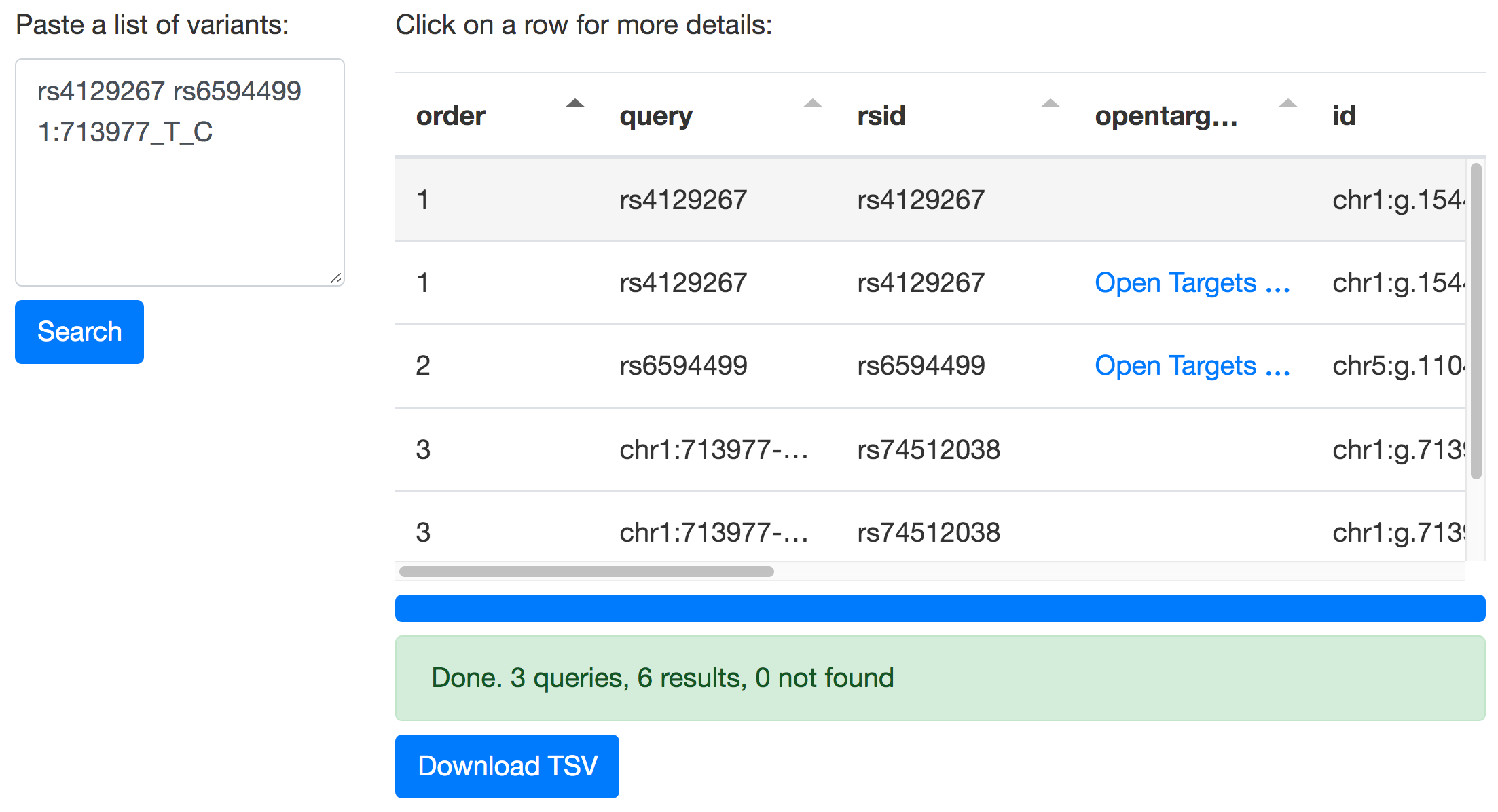
Make tidy variant tables with MyVariant.info and Tabulator
We can use the MyVariant.info API and Tabulator to create a web page for making tidy tables with genomic variants.
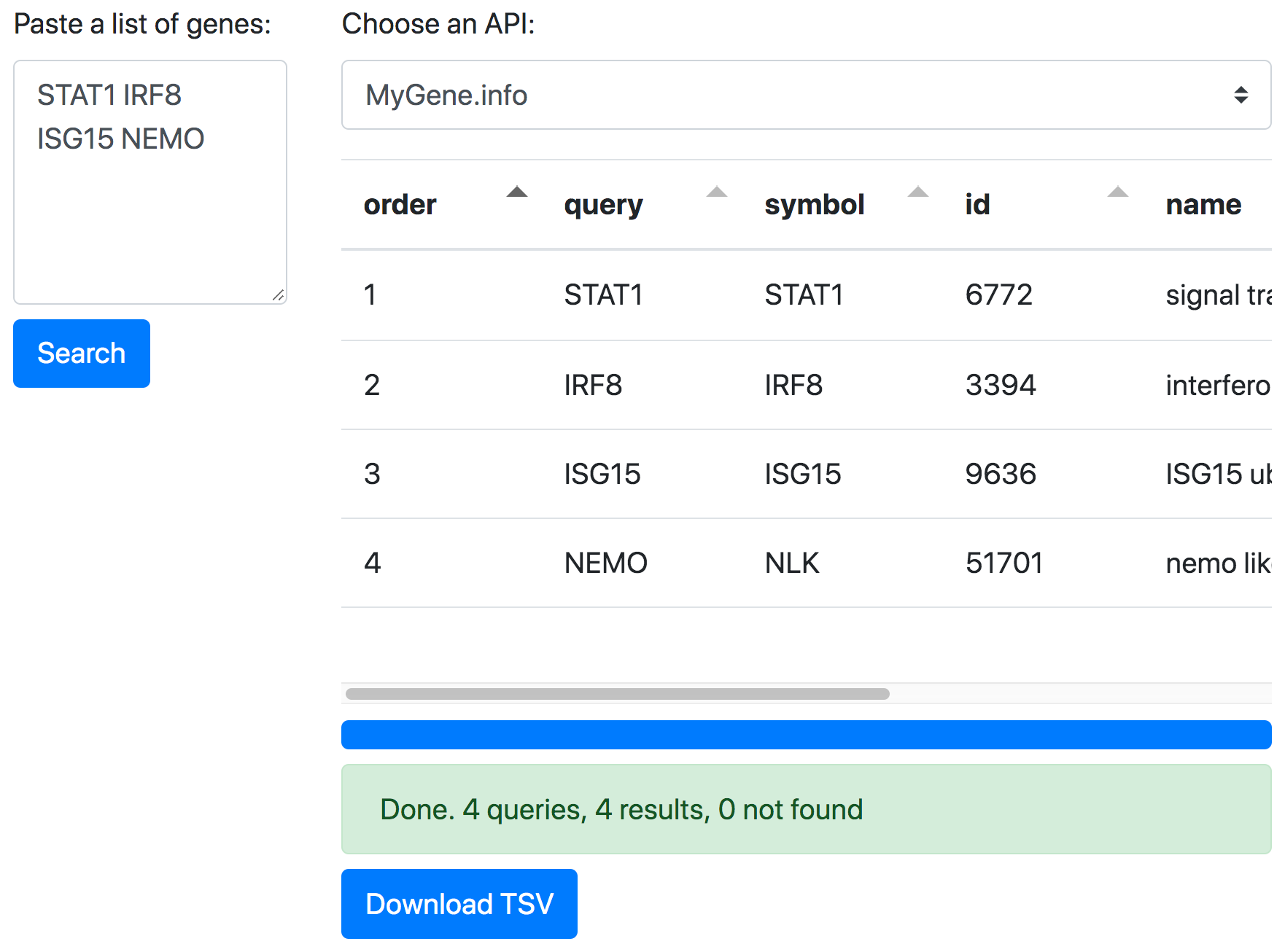
Make tidy gene tables with MyGene.info and Tabulator
Let’s use the MyGene.info API with the Tabulator JavaScript library by Oli Folkerd to create a simple web page for making tidy tables with information about genes. See how it works below, download the code, and try it yourself.
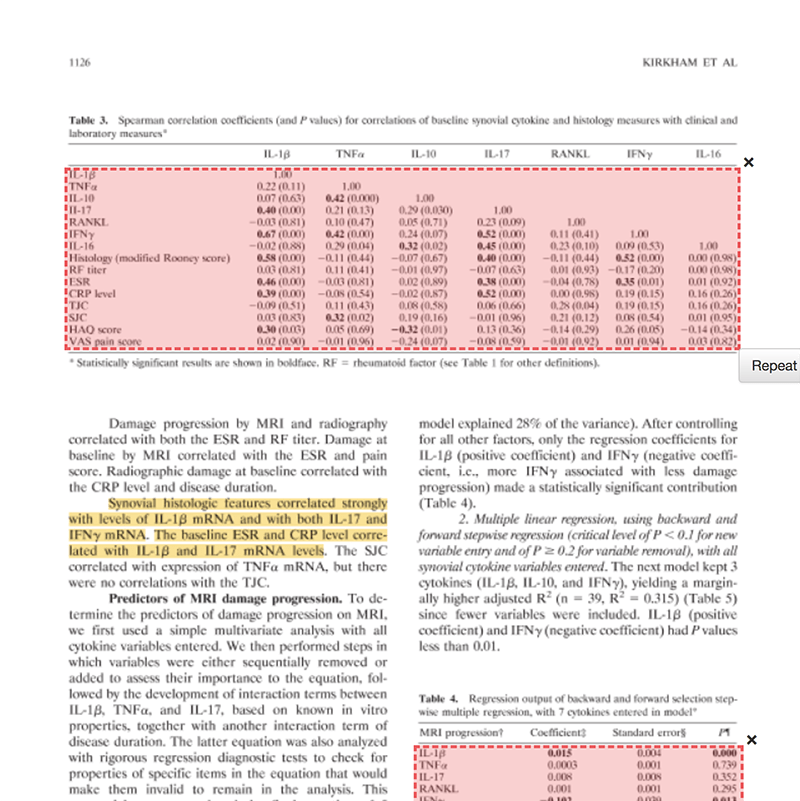
Extract data from a PDF file with Tabula
Kirkham et al. 2006 is a prospective 2-year study of 60 patients with rheumatoid arthritis (RA). It shows that “synovial membrane cytokine mRNA expression is predictive of joint damage progression in RA”. The PDF includes a few tables with data on cytokine measurements and correlations with joint damage. Here, we’ll use Tabula to extract data from tables in the PDF file. Then we’ll make figures with R.
Generate a large color palette with Colorgorical
Sometimes we need a lot of colors to represent all the categories in our data. We can use the httr and jsonlite packages to retrieve a list of colors from the Colorgorical website by Connor Gramazio.
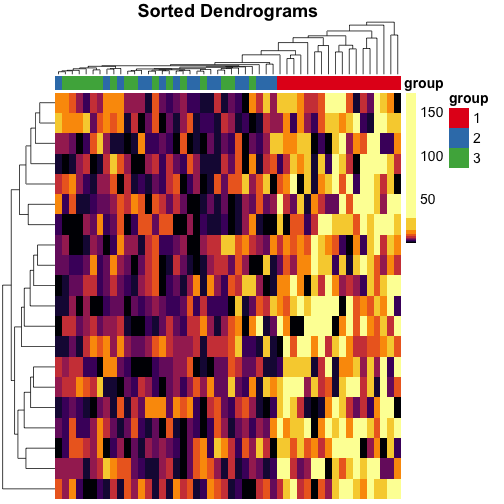
Make heatmaps in R with pheatmap
Here are a few tips for making heatmaps with the pheatmap R package by Raivo Kolde. We’ll use quantile color breaks, so each color represents an equal proportion of the data. We’ll also cluster the data with neatly sorted dendrograms, so it’s easy to see which samples are closely or distantly related.
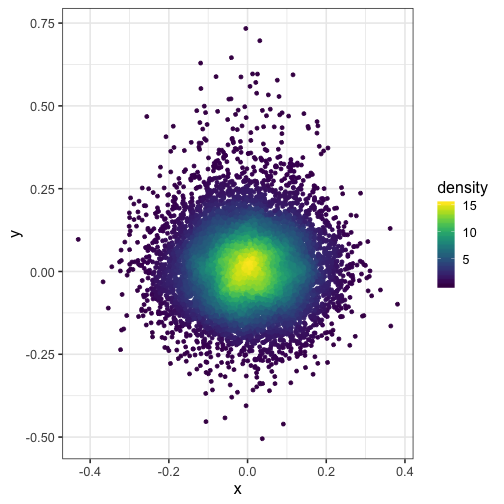
Color points by density with ggplot2
Here, we use the 2D kernel density estimation function from the MASS R package to to color points by density in a plot created with ggplot2. This helps us to see where most of the data points lie in a busy plot with many overplotted points.
Embed compressed data in HTML files
HTML is great for presenting rich text documents on the web. Javascript takes the web experience to the next level by allowing the content creator to develop scripts that run on the client-side in the visitors' web browsers. In this post, we’ll show a simple example of how we can embed arbitrary data into those scripts.
Build bioinformatics pipelines with Snakemake
Snakemake is a Pythonic variant of GNU Make. Recently, I learned how to use it to build and launch bioinformatics pipelines on an LSF cluster. However, I had trouble understanding the documentation for Snakemake. I like to learn by trying simple examples, so this post will walk you through a very simple pipeline step by step. If you already know how to use Snakemake, then you might be interested to copy my Snakefiles for RNA-seq data analysis here.
Determine if a transcription factor is bound to a genomic site with CENTIPEDE
I wrote a practical tutorial for how to use CENTIPEDE to determine if a transcription factor is bound to a site in the genome. The tutorial explains how to prepare appropriate input data and how to run the analysis. Please get in touch if you have any comments or suggestions.
Print bigWig data for each region in a BED file
I wrote a Bash script to call bigWigToBedGraph for each region in a BED file. You can quickly take a subset of bigWig data for regions of interest. In my particular case, I needed to get phastCons conservation scores for putative transcription factor binding sites.
Get human transcription factor target genes
I made a data package with human transcription factor target genes for use in R. It is a collection of data from three sources: TRED, ITFP, and ENCODE. I use them to test if the targets of a transcription factor are differentially expressed in my data. Also, I can test if a set of transcription factor target genes is enriched for some gene set of interest.
Quickly aggregate your data in R with data.table
In genomics data, we often have multiple measurements for each gene. Sometimes we want to aggregate those measurements with the mean, median, or sum. The data.table R package can do this quickly with large datasets.
In this note, we compute the average of multiple measurements for each gene in a gene expression matrix.
Make ribosomal RNA intervals for Picard CollectRnaSeqMetrics
Before you can use the CollectRnaSeqMetrics Picard tool, you must create a table of genomic intervals with the coordinates of all ribosomal genes in the genome. I wrote a bash script to prepare this ribosomal interval file from Gencode gene annotations.
Join multiple PLINK dosage files into one file
If you have multiple PLINK dosage files and would like to merge them into one file, this script might save you some time.
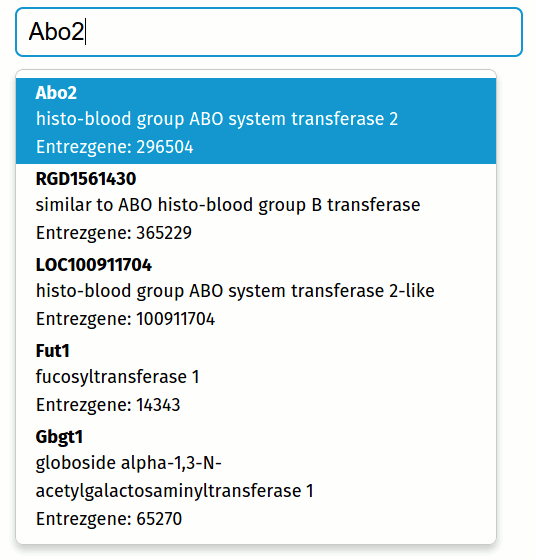
Autocomplete gene names with mygene.info and typeahead.js
We can use mygene.info with typeahead.js to autocomplete gene names and retrieve every annotation you can think of (GO, Kegg, Ensembl, position, homologs, etc.). Try typing your gene name in the interactive example in this post, and download my code.
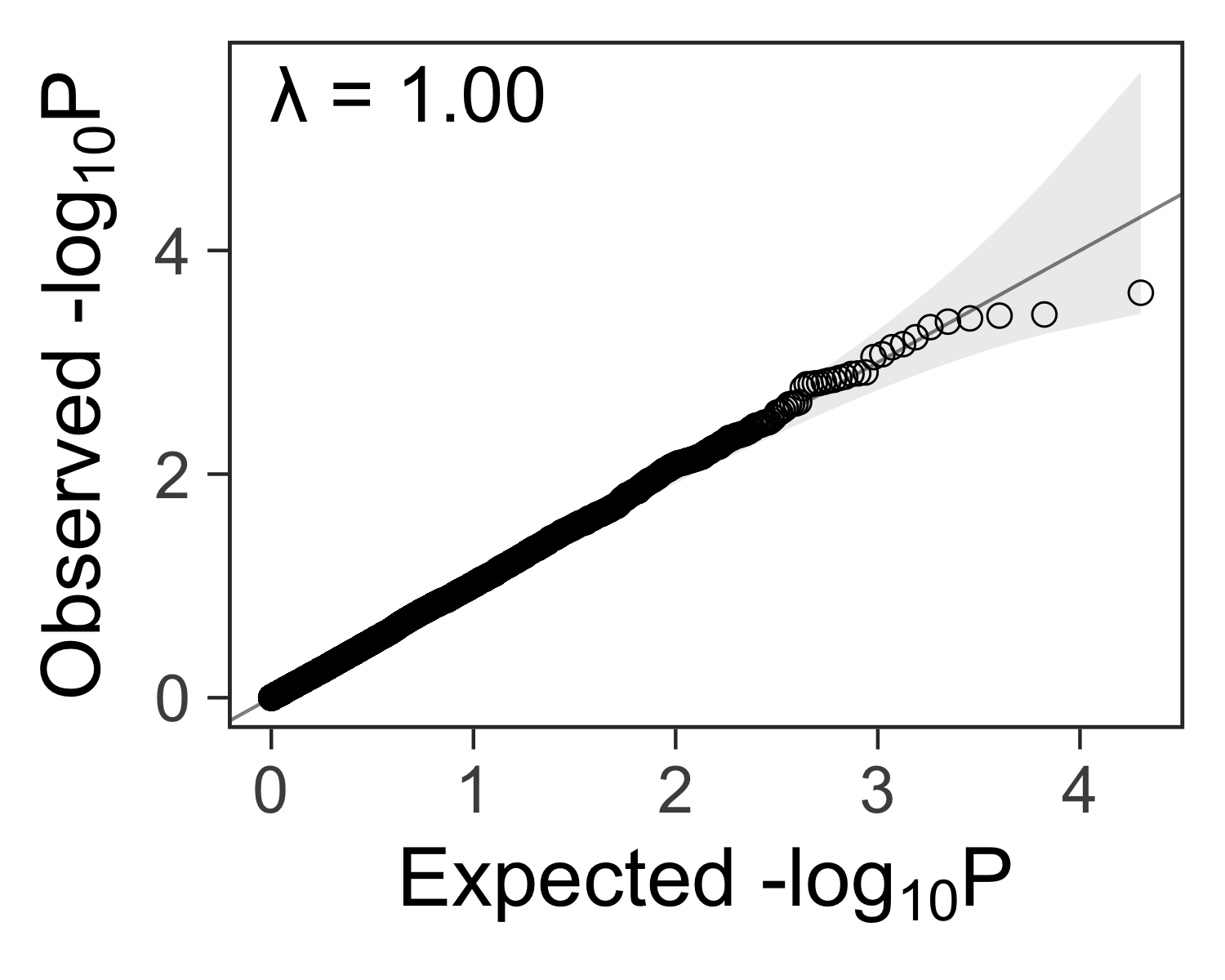
Create a quantile-quantile plot with ggplot2
After performing many tests for statistical significance, the next step is to check if any results are more extreme than we would expect by random chance. One way to do this is by comparing the distribution of p-values from our tests to the uniform distribution with a quantile-quantile (QQ) plot. Here’s a function to create such a plot with ggplot2.
How to ssh to a remote server without typing your password
Here are a few tips to use ssh more effectively. Login to your server using
public key encryption instead of typing a password. Use the ~/.ssh/config
file to create short and memorable aliases for your servers. Also, use aliases
to connect through a login server into a work server.
Count the number of coding base pairs in each Gencode gene
We can use Python to count the coding base pairs in each Gencode gene. Here, we report the base pair count by gene rather than by transcript. When we encounter different transcripts for the same gene with overlapping exons, we only count those base pairs once rather than multiple times.
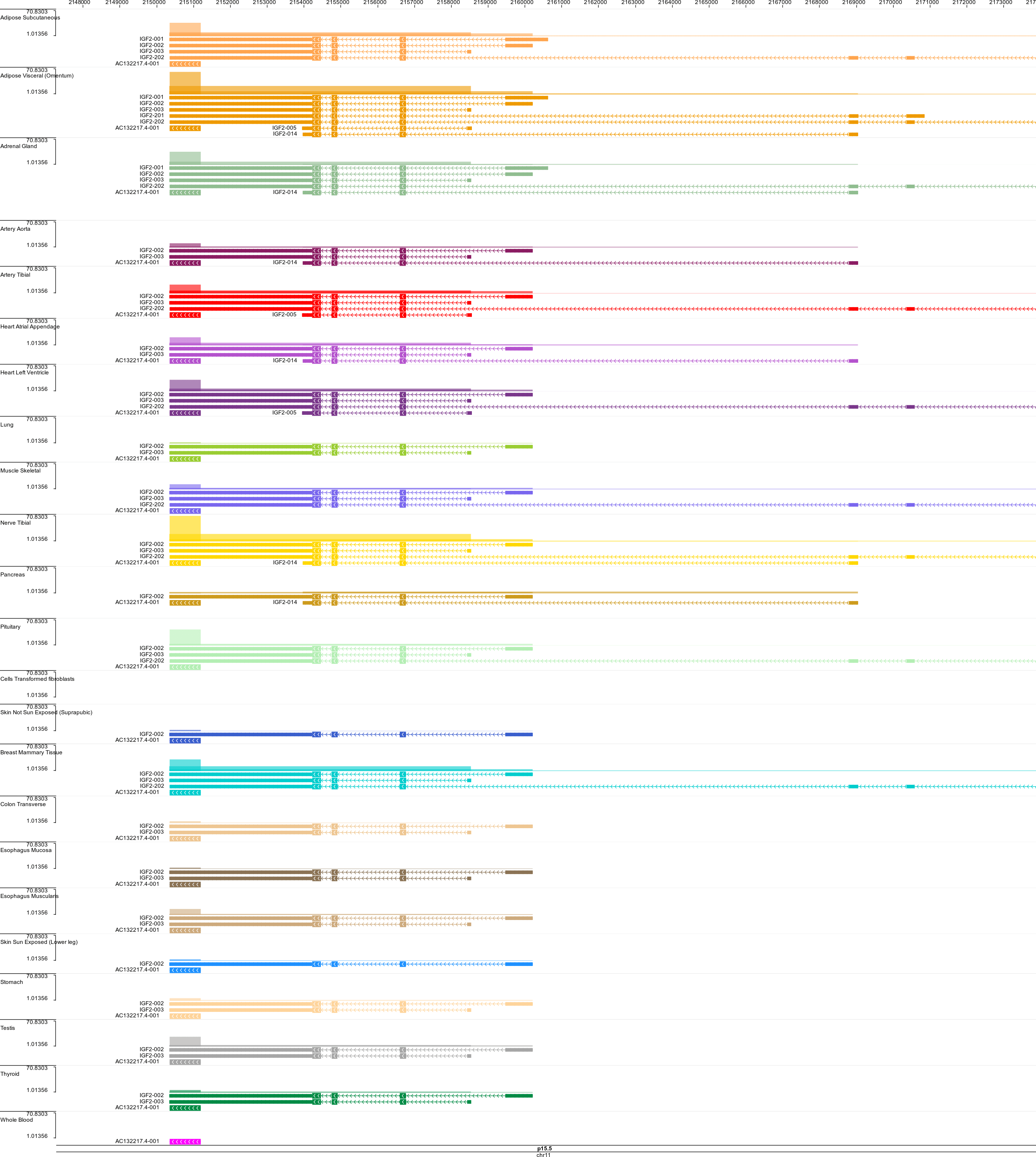
GTEx RNA-Seq Visualizations
I created three visualizations of RNA-Seq data from the GTEx project (version 2013-03-21). They’re powered by JBrowse, the WashU Epigenome Browser, and canvasXpress.
0-based and 1-based genomic intervals, overlap, and distance
Here, I describe two kinds of genomic intervals and include source code for testing overlap and calculating distance between intervals.