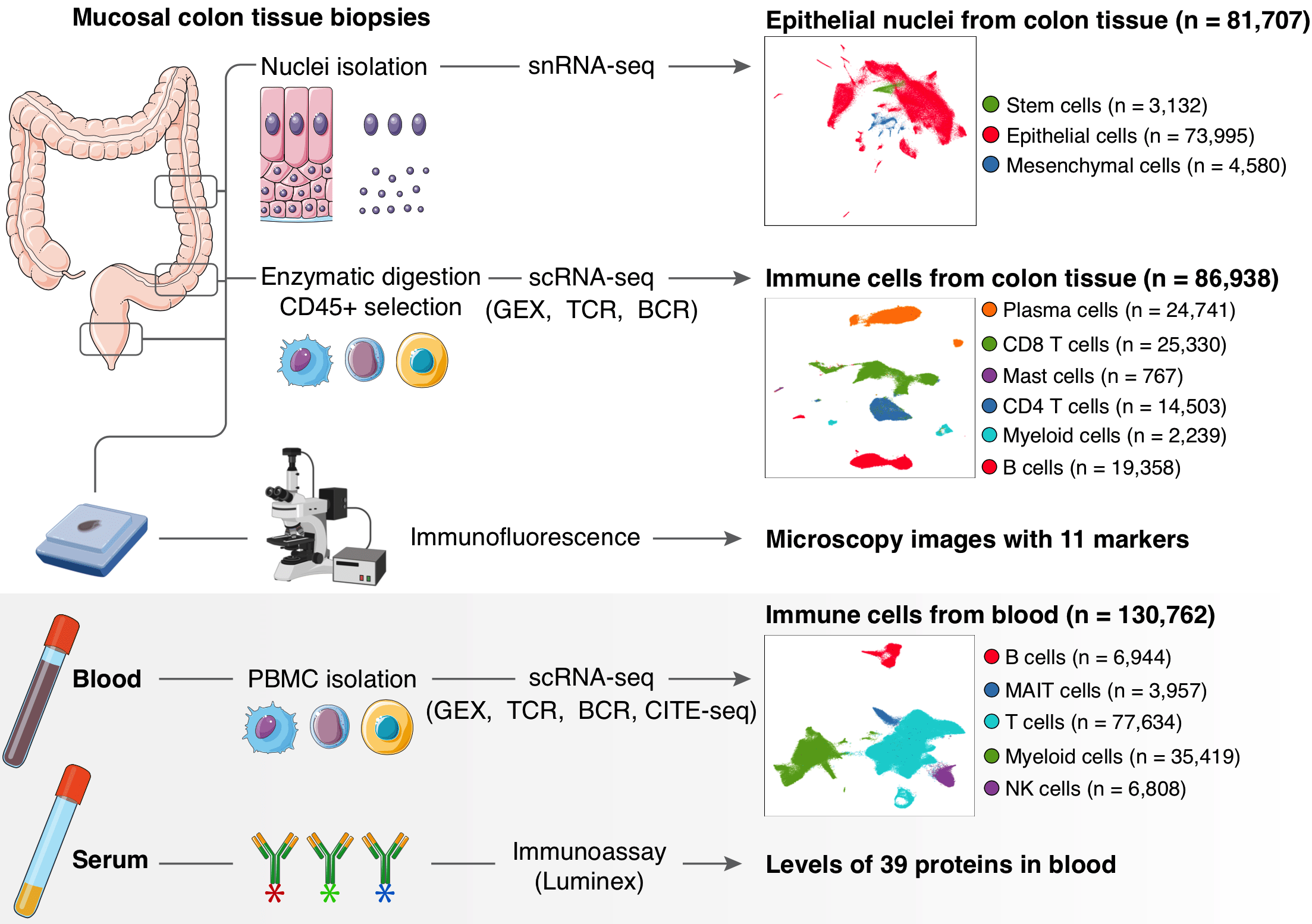
Altered interactions between circulating and tissue-resident CD8 T cells with the colonic mucosa define colitis associated with immune checkpoint inhibitors
Molly Fisher Thomas*, Kamil Slowikowski*, Kasidet Manakongtreecheep, Pritha Sen, Jessica Tantivit, Mazen Nasrallah, Neal P. Smith, Swetha Ramesh, Leyre Zubiri, Alice Tirard, Benjamin Y. Arnold, Linda T. Nieman, Jonathan H. Chen, Thomas Eisenhaure, Karin Pelka, Katherine H. Xu, Vjola Jorgji, Christopher J. Pinto, Tatyana Sharova, Rachel Glasser, Elaina PuiYee Chan, Ryan J. Sullivan, Hamed Khalili, Dejan Juric, Genevieve M. Boland, Michael Dougan, Nir Hacohen, Kerry L. Reynolds, Bo Li, Alexandra-Chloé Villani
bioRxiv, 2021. DOI: 10.1101/2021.09.17.460868
CUX1 and IκBζ (NFKBIZ) mediate the synergistic inflammatory response to TNF and IL-17A in stromal fibroblasts
Kamil Slowikowski*, Hung N. Nguyen*, Erika H. Noss, Daimon P. Simmons, Fumitaka Mizoguchi, Gerald F. M. Watts, Michael F. Gurish, Michael B. Brenner, and Soumya Raychaudhuri
PNAS, 2020. DOI: 10.1073/pnas.1912702117
Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single-cell transcriptomics and mass cytometry
Fan Zhang*, Kevin Wei*, Kamil Slowikowski*, Chamith Y. Fonseka*, Deepak A. Rao*, Stephen Kelly, Susan M. Goodman, Darren Tabechian, Laura B. Hughes, Karen Salomon-Escoto, Gerald F. M. Watts, A. Helena Jonsson, Javier Rangel-Moreno, Nida Meednu, Cristina Rozo, William Apruzzese, Thomas M. Eisenhaure, David J. Lieb, David L. Boyle, Arthur M. Mandelin II, Accelerating Medicines Partnership Rheumatoid Arthritis and Systemic Lupus Erythematosus (AMP RA/SLE) Consortium, Brendan F. Boyce, Edward DiCarlo, Ellen M. Gravallese, Peter K. Gregersen, Larry Moreland, Gary S. Firestein, Nir Hacohen, Chad Nusbaum, James A. Lederer, Harris Perlman, Costantino Pitzalis, Andrew Filer, V. Michael Holers, Vivian P. Bykerk, Laura T. Donlin, Jennifer H. Anolik, Michael B. Brenner & Soumya Raychaudhuri
Nature Immunology, 2019. DOI: 10.1038/s41590-019-0378-1
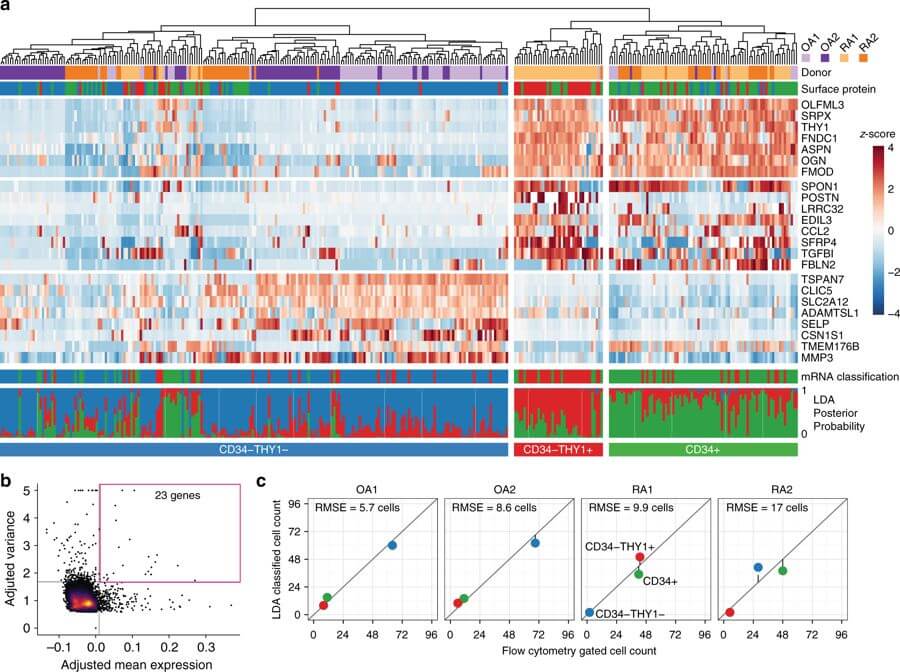
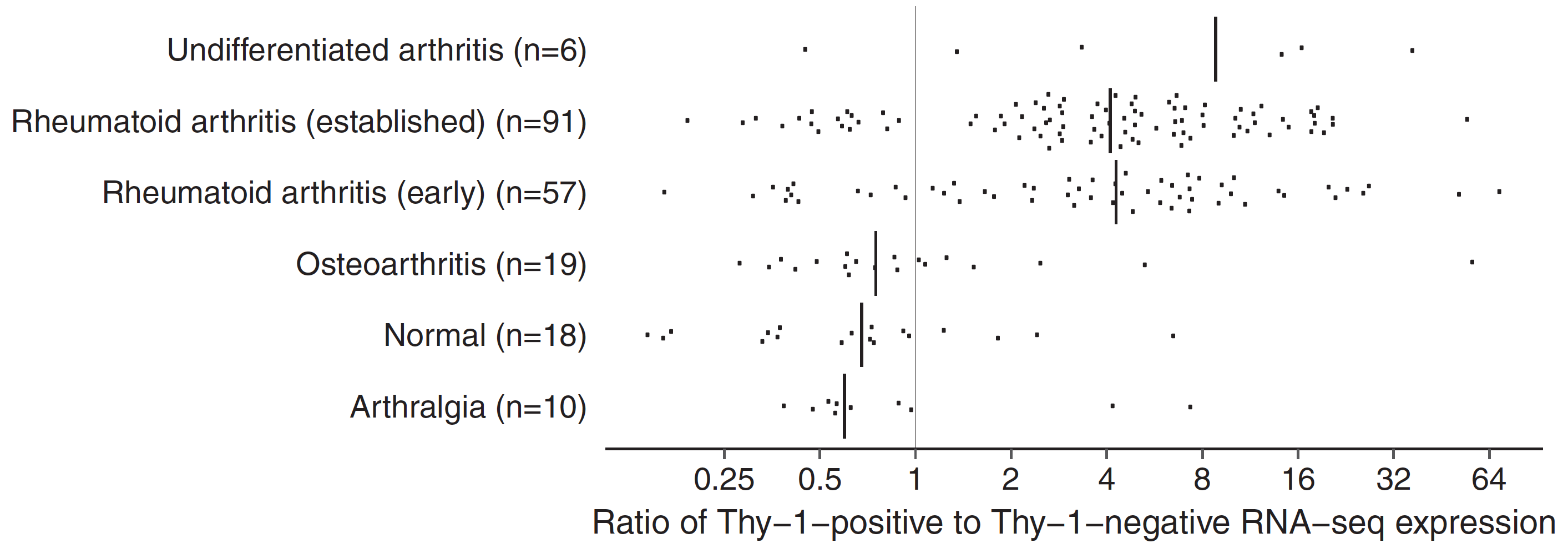
Functionally distinct disease-associated fibroblast subsets in rheumatoid arthritis
Fumitaka Mizoguchi*, Kamil Slowikowski*, Kevin Wei, Jennifer L. Marshall, Deepak A. Rao, Sook Kyung Chang, Hung N. Nguyen, Erika H. Noss, Jason D. Turner, Brandon E. Earp, Philip E. Blazar, John Wright, Barry P. Simmons, Laura T. Donlin, George D. Kalliolias, Susan M. Goodman, Vivian P. Bykerk, Lionel B. Ivashkiv, James A. Lederer, Nir Hacohen, Peter A. Nigrovic, Andrew Filer, Christopher D. Buckley, Soumya Raychaudhuri & Michael B. Brenner
Nature Communications, 2018. DOI: 10.1038/s41467-018-02892-y
Functional genomics of stromal cells in chronic inflammatory diseases
Kamil Slowikowski, Kevin Wei, Michael B. Brenner, Soumya Raychaudhuri
Current Opinion in Rheumatology, 2018. DOI: 10.1097/BOR.0000000000000455
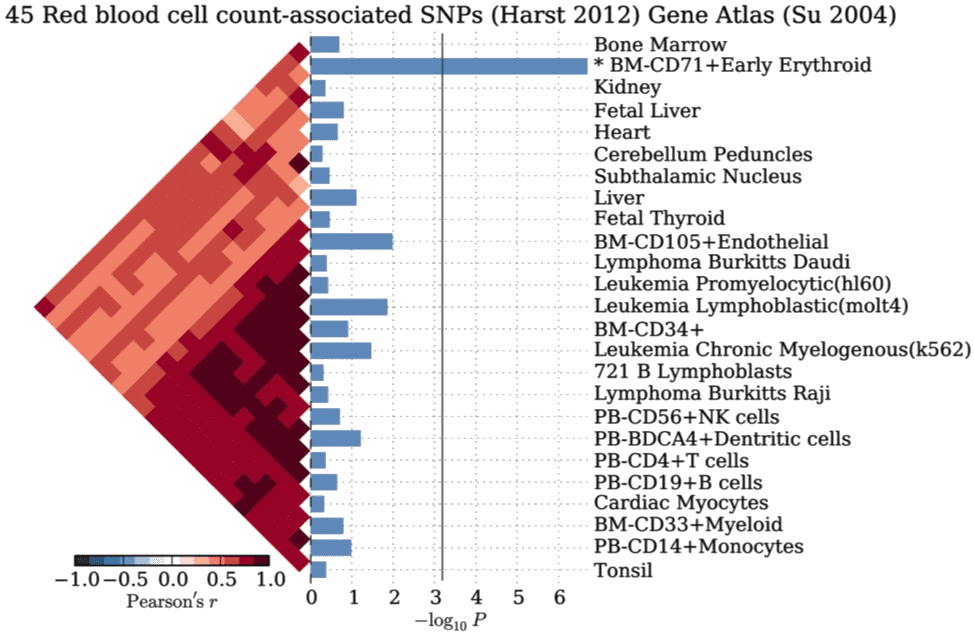
SNPSEA: an algorithm to identify cell types, tissues, and pathways affected by risk loci
Kamil Slowikowski, Xinli Hu, Soumya Raychaudhuri
Bioinformatics, 2014. DOI: 10.1093/bioinformatics/btu326