SNPSEA: an algorithm to identify cell types, tissues, and pathways affected by risk loci
Kamil Slowikowski, Xinli Hu, Soumya Raychaudhuri
Bioinformatics, 2014. DOI: 10.1093/bioinformatics/btu326
Abstract
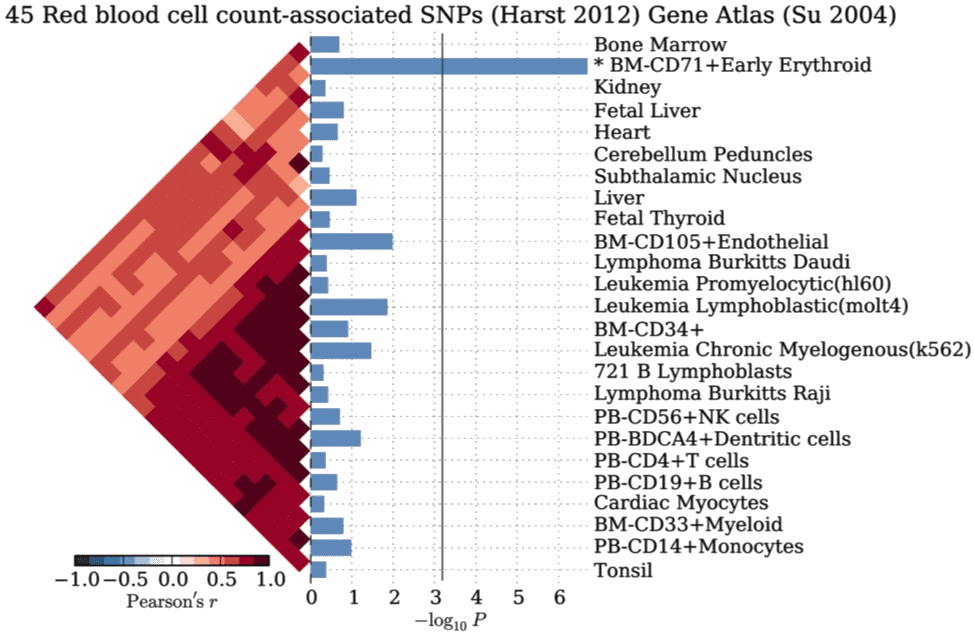
We created a fast, robust and general C++ implementation of a single-nucleotide polymorphism (SNP) set enrichment algorithm to identify cell types, tissues and pathways affected by risk loci. It tests trait-associated genomic loci for enrichment of specificity to conditions (cell types, tissues and pathways). We use a non-parametric statistical approach to compute empirical P-values by comparison with null SNP sets. As a proof of concept, we present novel applications of our method to four sets of genome-wide significant SNPs associated with red blood cell count, multiple sclerosis, celiac disease and HDL cholesterol.