SNPSEA #
SNPSEA is a single-nucleotide polymorphism (SNP) Set Enrichment Algorithm (SEA) to identify which cell types preferentially express the genes that are associated with a trait.
Input files:
- A list of genome-wide significant SNP identifiers (e.g.
rs42) from a genome-wide association study (GWAS) on your preferred trait. - A (NxM) matrix of gene expression values for all genes (N) across a large number (M) of cell types.
Example #
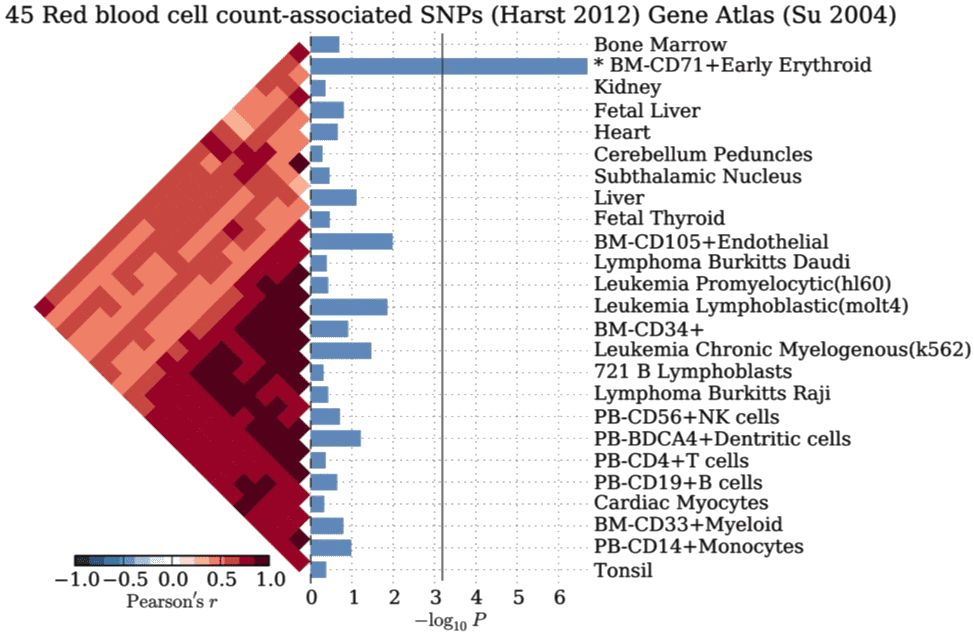
SNPSEA compares the trait-associated single-nucleotide polymorphisms (SNPs) to randomly sampled SNPs while accounting for linkage disequilibrium (LD). It’s implemented in C++ with executables available for macOS or Linux.
Reply by Email