Some grant agencies might require a table that lists all of your coauthors, departments, and dates for publications from the last few years. Making such a table can be a laborious task for academics who publish papers with a lot of coauthors. Here, we’ll download our publication records from NCBI and use R to automatically make a table of coauthors.
Download publications from NCBI in MEDLINE format #
Before we begin, set up a My Bibliography account at NCBI:
My Bibliography is a reference tool that helps you save your citations from PubMed or, if not found there, to manually upload a citations file, or to enter citation information using My Bibliography templates. My Bibliography provides a centralized place for your publications where citations are easily accessed, exported as a file, and made public to share with others.
Learn more at: https://www.ncbi.nlm.nih.gov/books/NBK53595/
Once My Bibliography is set up, we can get a text file in the MEDLINE format with all of our recent publications.
Go to this link:
https://www.ncbi.nlm.nih.gov/myncbi/collections/mybibliography/
It should look something like this:
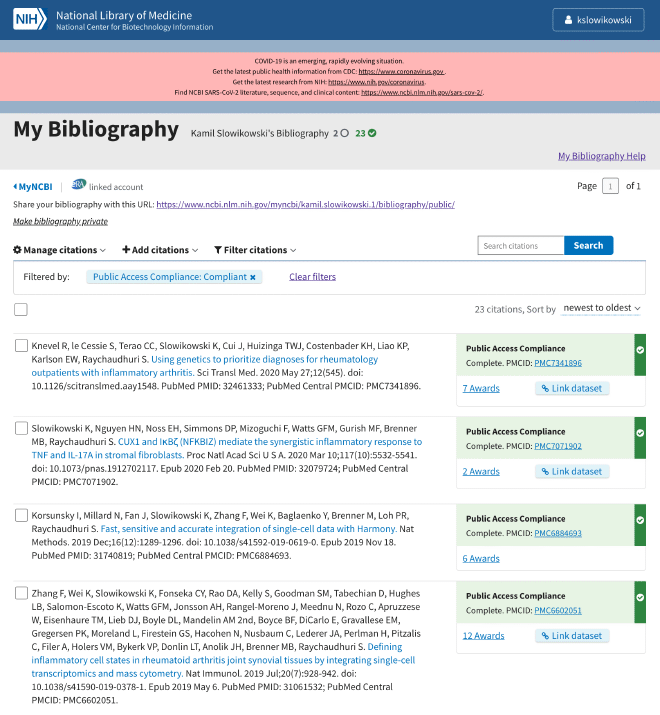
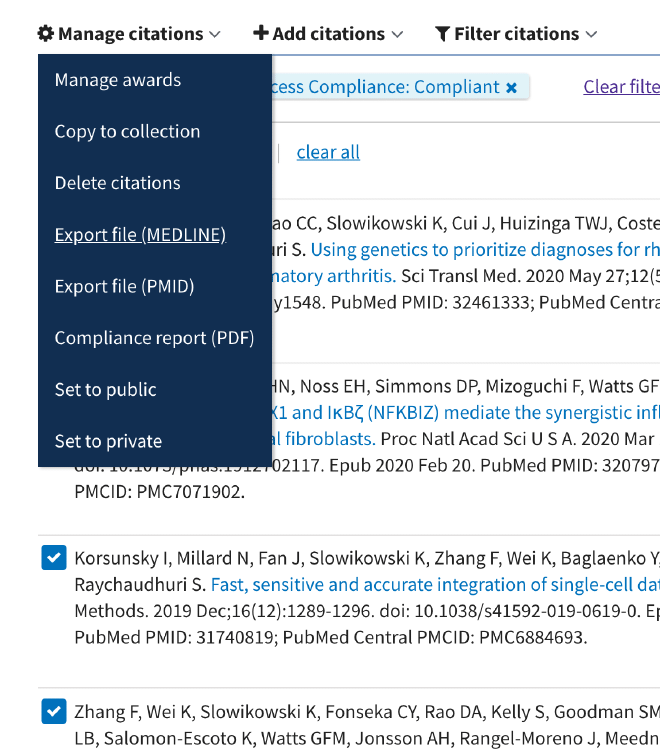
Click the checkboxes for the recent publications for which we want to get coauthors.
Click → Manage citations and → Export file (MEDLINE).
An example of the MEDLINE file #
Here is my file: medline.txt
Convert MEDLINE to TSV with an R script #
Run the following R script to convert the medline.txt file to a table with
coauthors in the format required by the grant agency.
Please feel free to copy and modify the code as you wish.
library(stringr)
library(tibble)
library(magrittr)
library(dplyr)
library(data.table)
# Read the text into a character vector of lines.
medline <- readLines("medline.txt")
# Discard lines with no content.
medline <- medline[nchar(medline) > 0]
# Concatentate all lines into a single string.
medline <- paste(medline, collapse = "\n")
# Unwrap long lines.
medline <- gsub("\n +", " ", medline)
# Split the string back into lines.
medline <- unlist(strsplit(medline, "\n"))
# Keep only some lines of interest.
lines <- medline[grepl("^(FAU|AD|DP)", medline)]
# Make a dataframe from the lines.
d <- str_split_fixed(lines, "- ", 2)
colnames(d) <- c("key", "value")
d <- as_tibble(d)
# Discard spaces from the "key" colun.
d$key <- str_replace_all(d$key, " ", "")
# Keep only the first affiliation for each author.
d <- d[with(d, c(key[-1] != key[-nrow(d)], TRUE)),]
# Assign an identifier to each publication.
d$id <- 0
d$id[d$key == "DP"] <- 1
d$id <- cumsum(d$id)
# For each publication, process the authors.
res <- rbindlist(lapply(sort(unique(d$id)), function(this_id) {
x <- d[d$id == this_id,]
# Assign an identifier to each author.
x$author_id <- 0
x$author_id[x$key == "FAU"] <- 1
x$author_id <- cumsum(x$author_id)
# Get the date for this publication.
this_date <- x$value[x$key == "DP"]
# Make a dataframe with columns: author, dept, date
x %>% filter(author_id > 0) %>%
group_by(author_id) %>%
summarize(
author = value[key == "FAU"],
dept = value[key == "AD"],
date = this_date,
.groups = "drop"
) %>%
select(-author_id)
})) %>% as_tibble
# Drop duplicated authors.
res <- res[!duplicated(res$author),]
# Drop extra departments.
res$dept <- str_split_fixed(res$dept, ";", 2)[,1]
# Write to file.
fwrite(res, "authors.tsv", sep = "\t")
An example of the TSV file #
Here’s what my generated file authors.tsv looks like:

Good luck with your grant application! I hope this note saves you some time.
Reply by Email